Annihilation of calcium oscillations
| Annihilation of calcium oscillations | |
|---|---|
| State dimension: | 1 |
| Differential states: | 4 |
| Discrete control functions: | 1 |
| Interior point equalities: | 4 |
The aim of the control problem is to identify strength and timing of inhibitor stimuli that lead to a phase singularity which annihilates intracellular calcium oscillations. This is formulated as an objective function that aims at minimizing the state deviation from a desired unstable steady state, integrated over time.
The mathematical equations form a small-scale ODE model. The interior point equality conditions fix the initial values of the differential states. The problem is, despite of its low dimension, very hard to solve, as the target state is unstable.
Contents
[hide]Biological motivation
Biological rhythms as impressing manifestations of self-organized dynamics associated with the phenomenon life have been of particular interest since quite a long time. Even before the mechanistic basis of certain biochemical oscillators was elucidated by molecular biology techniques, their investigation and the issue of perturbation by external stimuli has attracted much attention.
A calcium oscillator model describing intracellular calcium spiking in hepatocytes induced by an extracellular increase in adenosine triphosphate (ATP) concentration is described. The calcium signaling pathway is initiated via a receptor activated G-protein inducing the intracellular release of inositol triphoshate (IP3) by phospholipase C. The IP3 triggers the opening of endoplasmic reticulum and plasma membrane calcium channels and a subsequent inflow of calcium ions from intracellular and extracellular stores leading to transient calcium spikes.
As a source of external control a temporally varying concentration of an uncompetitive inhibitor of the PMCA ion pump is considered.
Mathematical formulation
For ![t \in [t_0, t_f]](https://mintoc.de/images/math/5/5/8/55823791d9100bcb5461801aff4f6edd.png) almost everywhere the mixed-integer optimal control problem is given by
almost everywhere the mixed-integer optimal control problem is given by
![\begin{array}{llcl}
\displaystyle \min_{x, w, w^{\mathrm{max}}} & & & {\int_{t_0}^{t_f} || x(\tau) - \tilde{x} ||_2^2 + p_1 w(\tau) \; \mathrm{d}\tau} \\[1.5ex]
\mbox{s.t.} & \dot{x}_0 & = & k_1 + k_2 x_0 - \frac{k_3 x_0 x_1}{x_0 + K_4} - \frac{k_5 x_0 x_2}{x_0 + K_6} \\
& \dot{x}_1 & = & k_7 x_0 - \frac{k_8 x_1}{x_1 + K_9} \\
& \dot{x}_2 & = & \frac{k_{10} x_1 x_2 x_3}{x_3 + K_{11}} + k_{12} x_1 + k_{13} x_0 - \frac{k_{14} x_2}{{w} x_2 + K_{15}} - \frac{k_{16} x_2}{x_2 + K_{17}} + \frac{x_3}{10} \\
& \dot{x}_3 & = & - \frac{k_{10} x_1 x_2 x_3}{x_3 + K_{11}} + \frac{k_{16} x_2}{x_2 + K_{17}} - \frac{x_3}{10} \\[1.5ex]
& x(0) &=& (0.03966, 1.09799, 0.00142, 1.65431)^T, \\
& x(t) & \ge & 0.0, \\
& w(t) &\in& \{0, w^{\mathrm{max}}\}, \\
& w^{\mathrm{max}} & \ge & 1.1, \\
& w^{\mathrm{max}} & \le & 1.3.
\end{array}](https://mintoc.de/images/math/b/7/a/b7a2070f63c76a13be7fe40c97d61362.png)
Here the differential states 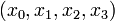 describe concentrations of activated G-proteins, active phospholipase C, intracellular calcium, and intra-ER calcium, respectively.
describe concentrations of activated G-proteins, active phospholipase C, intracellular calcium, and intra-ER calcium, respectively.
Modeling details including a comprehensive discussion of parameter values and the dynamical behavior observed in simulations with a comparison to experimental observations can be found in <bibref>Kummer2000</bibref>. In the given equations that stem from <bibref>Lebiedz2005</bibref>, the model is identical to the one derived there, except for an additional first-order leakage flow of calcium from the ER back to the cytoplasm, which is modeled by 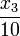 in equations 3 and 4. It reproduces well experimental observations of cytoplasmic calcium oscillations as well as bursting behavior and in particular the frequency encoding of the triggering stimulus strength, which is a well known mechanism for signal processing in cell biology.
in equations 3 and 4. It reproduces well experimental observations of cytoplasmic calcium oscillations as well as bursting behavior and in particular the frequency encoding of the triggering stimulus strength, which is a well known mechanism for signal processing in cell biology.
Parameters
These fixed values are used within the model.
![\begin{array}{rcl}
[t_0, t_f] &=& [0, 22],\\
k_1 &=& 0.09, \\
k_2 &=& 2.30066, \\
k_3 &=& 0.64, \\
K_4 &=& 0.19, \\
k_5 &=& 4.88, \\
K_6 &=& 1.18, \\
k_7 &=& 2.08, \\
k_8 &=& 32.24, \\
K_9 &=& 29.09, \\
k_{10} &=& 5.0, \\
K_{11} &=& 2.67, \\
k_{12} &=& 0.7, \\
k_{13} &=& 13.58, \\
k_{14} &=& 153.0, \\
K_{15} &=& 0.16, \\
k_{16} &=& 4.85, \\
K_{17} &=& 0.05, \\
p_1 &=& 100, \\
\tilde{x}_0 &=& 6.78677, \\
\tilde{x}_1 &=& 22.65836, \\
\tilde{x}_2 &=& 0.384306, \\
\tilde{x}_3 &=& 0.28977.
\end{array}](https://mintoc.de/images/math/a/b/8/ab8916d401588f2eb07c1ce89e5ef457.png)
Reference Solutions
- Reference solution plots
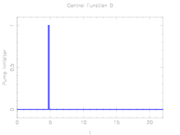
The depicted optimal solution consists of a stimulus of 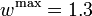 and a timing given by the stage lengths 4.6947115, 0.1491038, and 17.1561845. The optimal objective function value is 1.610654. As can be seen from the additional plots, this solution is extremely unstable. A small perturbation in the control, or simply rounding errors on a longer time horizon lead to a transition back to the stable limit-cycle oscillations.
and a timing given by the stage lengths 4.6947115, 0.1491038, and 17.1561845. The optimal objective function value is 1.610654. As can be seen from the additional plots, this solution is extremely unstable. A small perturbation in the control, or simply rounding errors on a longer time horizon lead to a transition back to the stable limit-cycle oscillations.
Optimization
The determination of the stimulus by means of optimization is quite hard for two reasons. First, the unstable target steady-state. Only a stable all-at-once algorithm such as multiple shooting or collocation can be applied successfully.
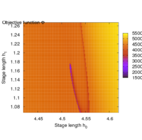
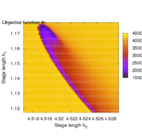
Second, the objective landscape of the problem in switching time formulation (this is, for a fixed stimulus strength and modifying only beginning and length of the stimulus) is quite nasty, as the following visualizations obtained by simulation show.
- Objective landscape simulation plots
Source Code
C
The differential equations in C code:
double myu = p[17] * (1.0 + u[0]* ( p[19] - 1.0) ) ; rhs[0] = p[0] + p[1]*xd[0] - p[2]*xd[0]*xd[1]/(xd[0]+p[3]) - p[4]*xd[0]*xd[2]/(xd[0]+p[5]); rhs[1] = p[6]*xd[0] - p[7]*xd[1]/(xd[1]+p[8]); rhs[2] = p[9]*xd[1]*xd[2]*xd[3]/(xd[3]+p[10]) + p[18]*p[11]*xd[1] + p[12]*xd[0] - p[13]*xd[2]/(myu*xd[2]+p[14]) - p[15]*xd[2]/(xd[2]+p[16]) + 0.1*xd[3]; rhs[3] = -p[9]*xd[1]*xd[2]*xd[3]/(xd[3]+p[10]) + p[15]*xd[2]/(xd[2]+p[16]) - 0.1*xd[3];
Variants
References
<bibreferences/>